| Here we give an example of QSP model for RG7652, published in Gadkar et al., CPT Pharmacometrics Syst. Pharmacol. 3, (2014) and translated in Mlxtran. A Monolix project estimating this model on phase I clinical data extracted from the plots from the same publication can be downloaded here.
Compared to PK/PD models, QSP models incorporate more mechanistic details and model entities which have not been measured experimentally. By putting more emphasis on the biological relevance, QSP models are believed to be more capable of extrapolating from preclinical to clinical or from healthy volunteers to patients.
The poster given on the right compares this QSP model to a typical PK/PD model |
|
Anti-PCSK9 mAb for cholesterol lowering
RG7652 [1] is a fully human monoclonal antibody antagonizing PSCK9 activity. It is developed as cholesterol (LDLc) lowering therapy and can be given in patients already treated with statins.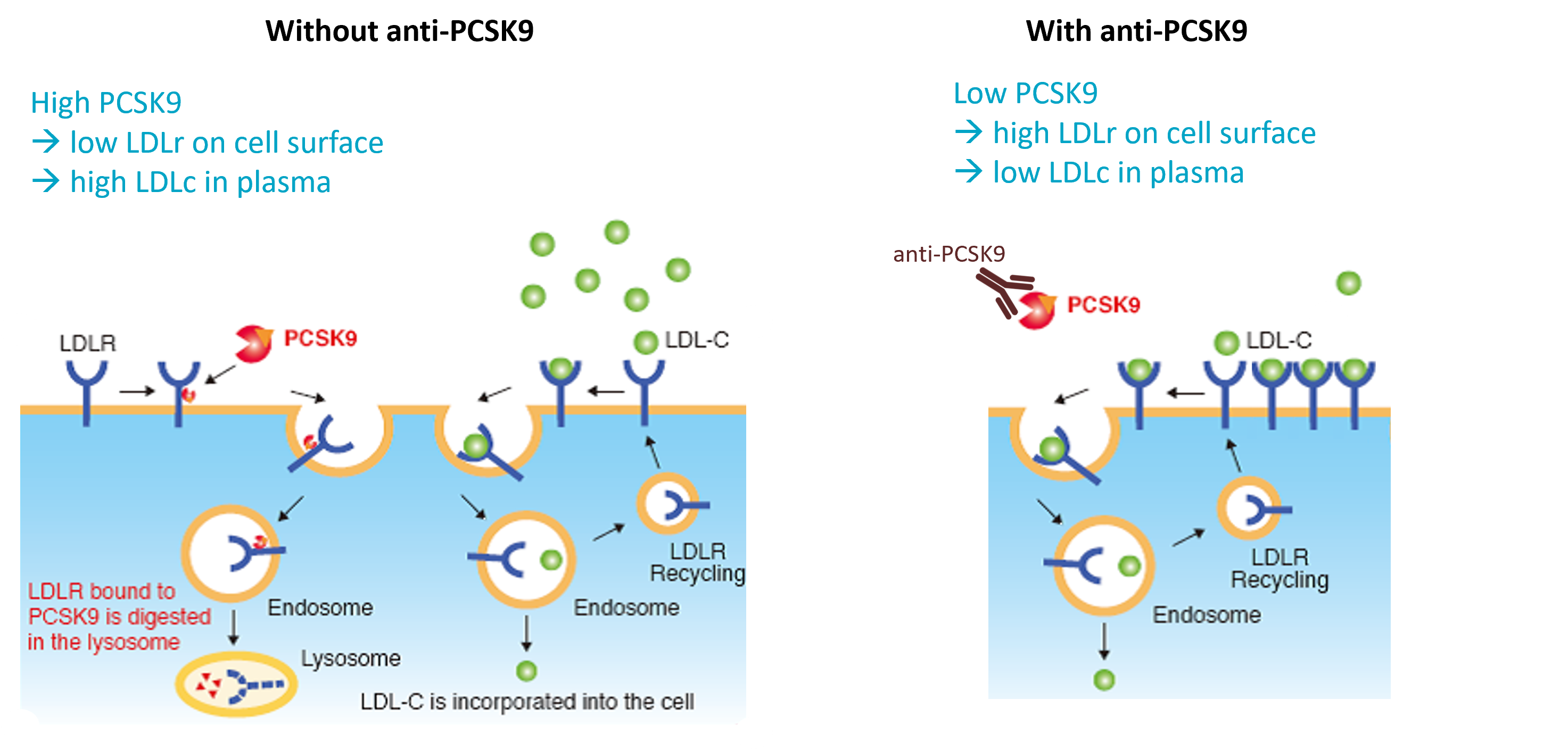
QSP model
The structural model (full text given below) is composed of six ODEs and 21 parameters. The modeled entities are:
- L: Drug
- P: Drug-PCSK9 complex
- PCSK9: Circulating PCSK9
- LDLr: LDL receptor
- LDLc: LDL cholesterol
- HepCh: Hepatic cholesterol
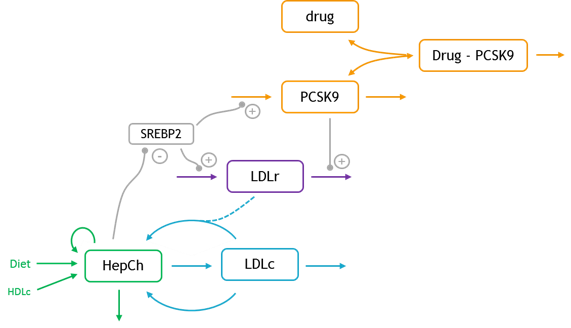
The model allows to investigate several questions by simulation, such as statin effect or the response of patients with familial hypercholesterolemia (FH).
Simulating statin effect
Statins induce SREBP2 which increases PCSK9 and LDLr levels (via increased synthesis), increased LDLr levels and may also decrease hepatic cholesterol synthesis. Thus the effect of statin can be implemented with:
- ksynPCSK9 increased, kdegPCSK9 same (but LDLr increased) => PCSK9 increased
- ksynLDLr increased, kdegLDLr same (but PCSK9 increased) => LDLr increased
- ksynLDLc same, kdegLDLc same but LDLr increased => LDLc decreased
Response of patients with familial hypercholesterolemia
These patients have a low concentrations of LDLr receptor due to mutations.
Full structural model
[LONGITUDINAL]
input = {ka, V, Cl, kint, KD, S_diet, f_diet_abs,S_HepCh_syn, f_HepCh_statin, k_HepCh_loss, k_LDL_syn, Ch_LDLparticle, k_LDLc_clr1, f_LDLcHep_clr, k_LDLc_clr2, k_HDLc_clr, Vblood, LDLr0, k_LDLr_deg, k_PCSK9_deg, k_PCSK9_syn}
PK:
MW = 150
depot(target=L, ka, p=1/V * 1000/MW)
; dose in mg
; L in nmol/L
; V in L
EQUATION:
odeType=stiff
t_0 = -100
L_0 = 0
P_0 = 0
HepCh_0 = 6000
LDLc_0 = 160
LDLr_0 = 1
PCSK9_0 = k_PCSK9_syn/k_PCSK9_deg
k_LDLr_syn = LDLr0 * k_LDLr_deg
HDLc = 50
kel = Cl/V
kon = 10
koff = KD*kon
; ======== effect of HepCh on PCSK9
maxSREBP2level = 2
minSREBP2level = 0
HepChref = 6000
expT=3.97865*3^-0.995 ; This is fitted to give 99% output at the extreme of the range specified
f_HepCh_reg=( minSREBP2level - maxSREBP2level ) * (HepCh^expT/(HepCh^expT+HepChref^expT)) + maxSREBP2level
SREBP2 = f_HepCh_reg
; ======== effect of PCSK9 on LDLr degradation
nM2ngPermL=74
pcsk9_on_LDLr = 0.75
Baselinepcsk9 = 281.94
pcsk9_on_LDLr_range = 650
expT=3.97865*2^-0.995
Xmod=10^( 2*(PCSK9*nM2ngPermL-(Baselinepcsk9-pcsk9_on_LDLr_range/2))/pcsk9_on_LDLr_range -1 )
Xmodic50=10^( 2*(Baselinepcsk9-(Baselinepcsk9-pcsk9_on_LDLr_range/2))/pcsk9_on_LDLr_range -1 )
f_PCSK9_degLDLr = 2*pcsk9_on_LDLr * (Xmod^expT/(Xmod^expT+Xmodic50^expT)) + (1-pcsk9_on_LDLr)
; ======== effect of SREBL2 on LDLr
VmaxUpLDLr=3
KmUpLDLr=1.5
VmaxDownLDLr=0.7
KmDownLDLr=0.5
if SREBP2>1
f_SREBP2_regLDLr = 1+ (VmaxUpLDLr-1) * (SREBP2-1)/((SREBP2-1)+(KmUpLDLr-1))
else
f_SREBP2_regLDLr = 1 - VmaxDownLDLr * (1-SREBP2)/((1-SREBP2)+KmDownLDLr)
end
; ======== effect of SREBL2 on PCSK9
VmaxUpPCSK9=2
KmUpPCSK9=1.5
VmaxDownPCSK9=0.7
KmDownPCSK9=0.5
if SREBP2>1
f_SREBP2_regPCSK9 = 1+ (VmaxUpPCSK9-1) * (SREBP2-1)/((SREBP2-1)+(KmUpPCSK9-1))
else
f_SREBP2_regPCSK9 = 1 - VmaxDownPCSK9 * (1-SREBP2)/((1-SREBP2)+KmDownPCSK9)
end
; ========= ODEs
ddt_HepCh = f_diet_abs * S_diet + f_HepCh_statin * S_HepCh_syn - k_HepCh_loss * HepCh - k_LDL_syn * Ch_LDLparticle * Vblood * HepCh + f_LDLcHep_clr * k_LDLc_clr1 *Vblood * LDLc * LDLr + f_LDLcHep_clr * k_LDLc_clr2 * Vblood * LDLc + f_LDLcHep_clr * k_HDLc_clr * Vblood * HDLc
ddt_LDLc = k_LDL_syn * Ch_LDLparticle * HepCh - k_LDLc_clr1 * LDLc * LDLr - k_LDLc_clr2 * LDLc
ddt_LDLr = k_LDLr_syn * f_SREBP2_regLDLr - k_LDLr_deg * LDLr * f_PCSK9_degLDLr
ddt_PCSK9 = k_PCSK9_syn * f_SREBP2_regPCSK9 - k_PCSK9_deg * PCSK9 - kon*L*PCSK9 + koff*P
ddt_P = kon*L*PCSK9 - koff*P - kint*P
ddt_L = -kel*L - kon*L*PCSK9 + koff*P
Ltot = L+P
Rtot = PCSK9+P
LDLc_change = LDLc/160*100
PCSK9_ngmL = PCSK9*74 ; from nmol/L to ng/mL
LDLc_mgdL = LDLc
OUTPUT:
output={Ltot, Rtot, LDLc_change}
