We here present the system of equation corresponding to the original (full) TMDD model, as well as the model behavior.
Equations
The first TMDD model has been proposed in Mager & Jusko (2001), JPP 28(6). The equations read:
with L the concentration of the ligand in plasma, R the concentration of the target/receptor, and P the concentration of the complex. V is the volume of the central compartment for the ligand, kel the linear elimination rate for the ligand, kon the binding rate, koff the dissociation rate, ksyn the synthesis rate for the target/receptor, kdeg its degradation rate, and kint the degradation rate of the complex. In(t) represents the input function, corresponding to the input rate (amount per unit time) of the ligand into the central compartment due to the ligand administration.
In the library model file, a slightly different parameterization is used using the steady-state initial receptor concentration (which replaces kdeg in the list of parameters), and
the dissociation constant (which replaces koff in the list of parameters). This permits to better separate the effect of each parameter.
If in addition the ligand can distribute to peripheral tissues, a second compartment can be added, leading to:
with A the amount of ligand in peripheral tissues, k12 the rate of transfert from central to peripheral, and k21 the rate in the opposite direction.
The main assumptions of this model are the following (Gibiansky et al. PAGE 2011 talk):
- the binding of the ligand to the target is simple, not cooperative
- binding occurs only in the central compartment
- target and complex do not diffuse to peripheral compartments
- internalized ligand-receptor complexes are not recycled
Model properties
To better understand the properties of such a model, we propose to explore the typical concentration-time curves of this model and the influence of the parameter values. We will focus on the concentration-time curve of the free ligand after a bolus dose, while the other species can be investigated by the reader using the provided Mlxplore project files. The vignettes below explicit the impact of each parameter on the typical free ligand concentration-time curve:
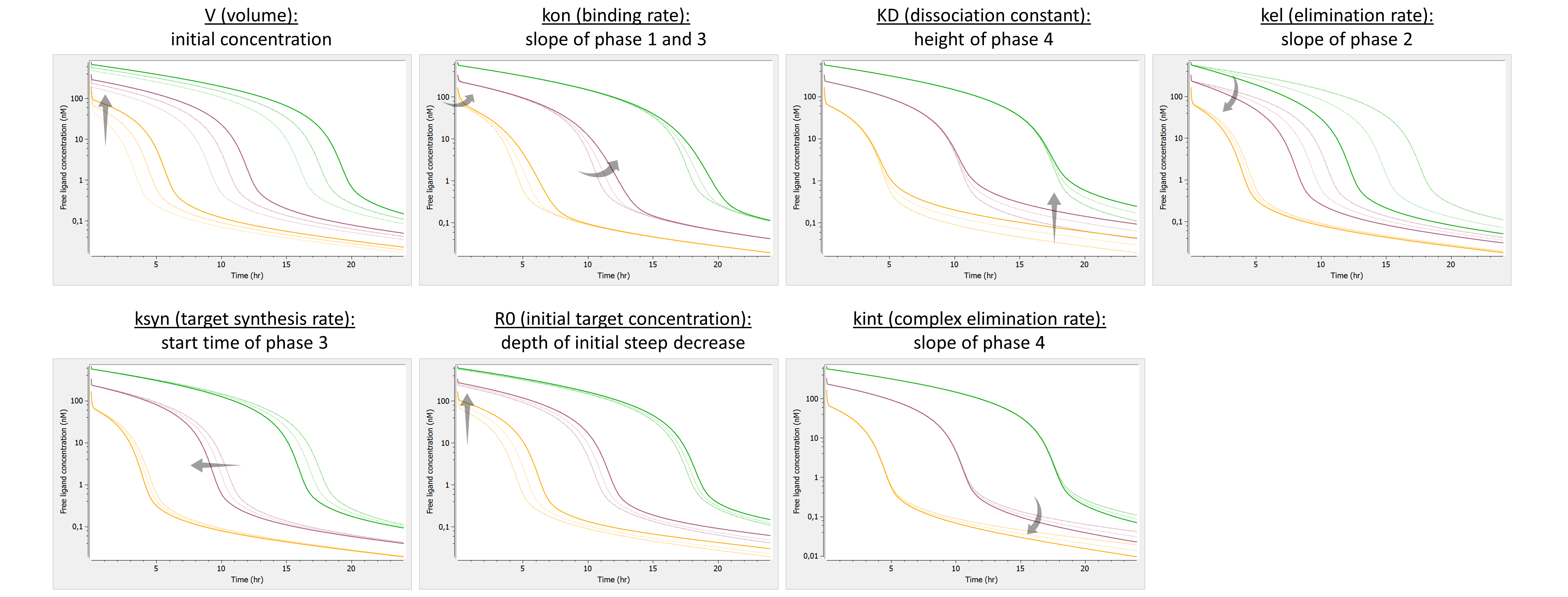
The effects are summarized below:
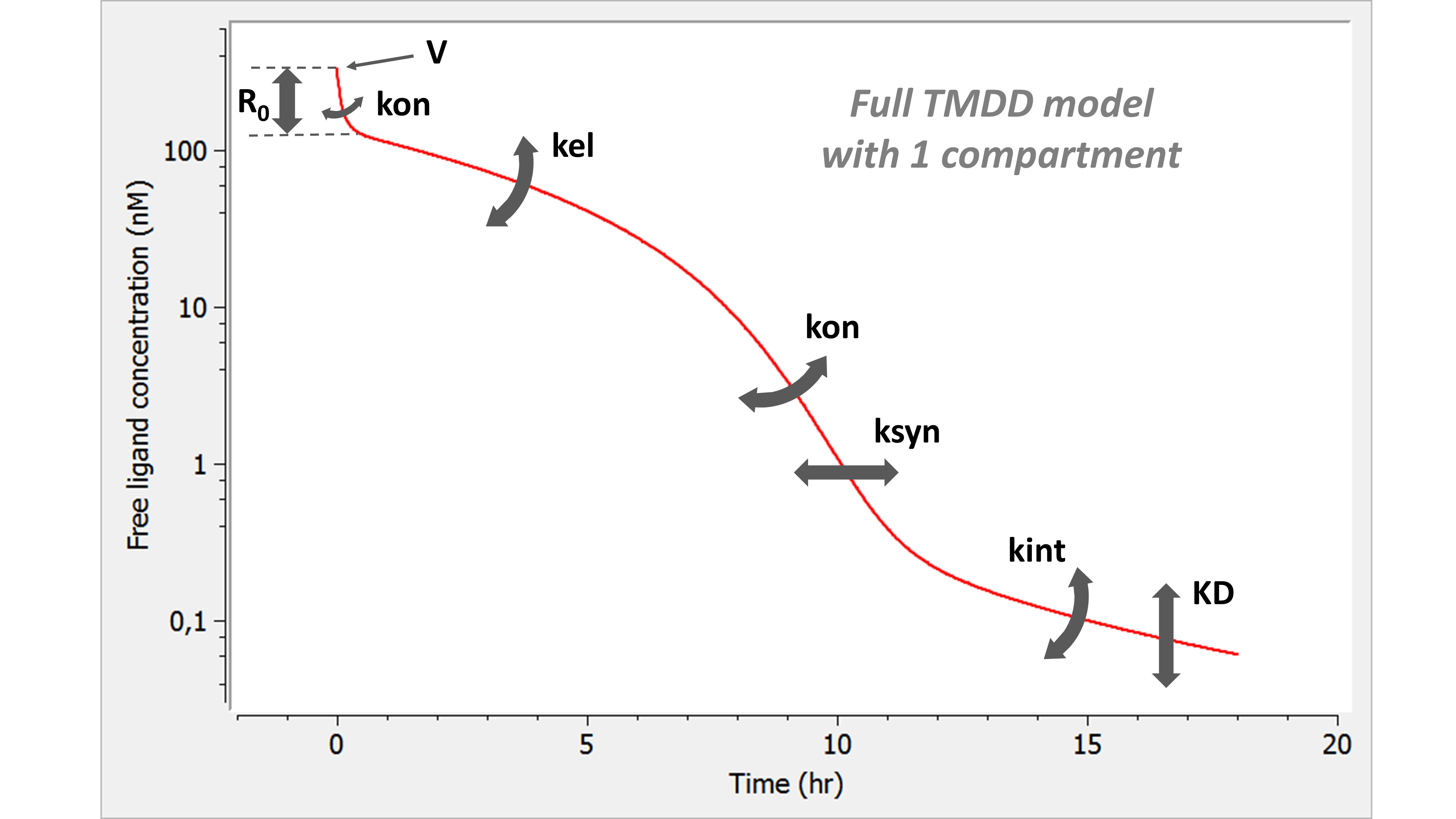 For every phase of the concentration-time curve, there exists a parameter to modify it. This is in contrast with the various approximations that lack flexibility for one or several phases of the curve.
For every phase of the concentration-time curve, there exists a parameter to modify it. This is in contrast with the various approximations that lack flexibility for one or several phases of the curve.
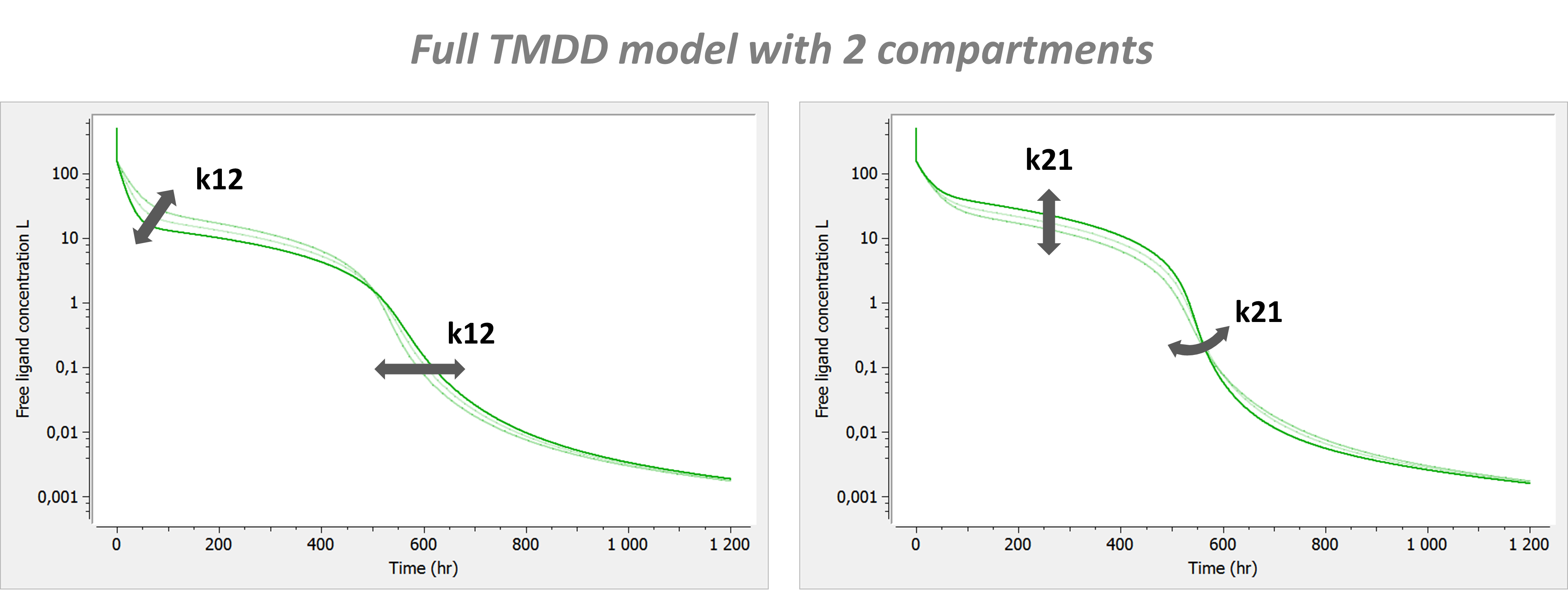
Model with 2 compartments
If a second compartment is added, the shape is modified in the following way. Note that kon has been chosen large (very steep phase 1), to better focus on phase 2, where k12 and k21 have their main effect.
Click here to go back to main TMDD page.